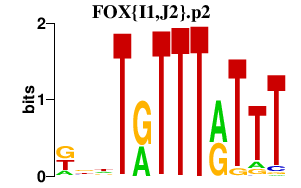
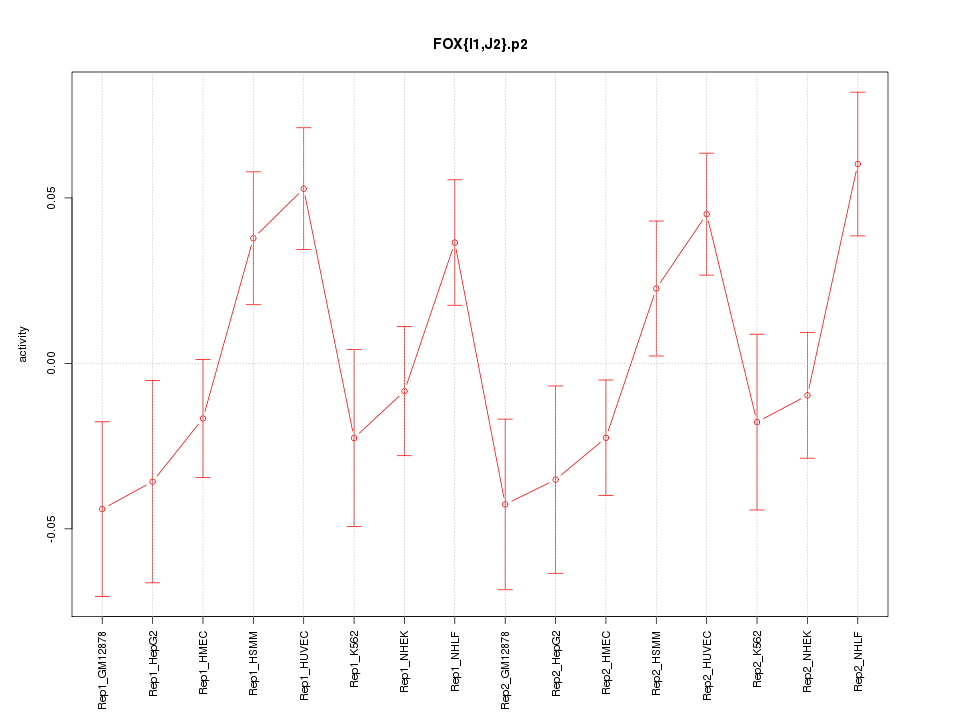
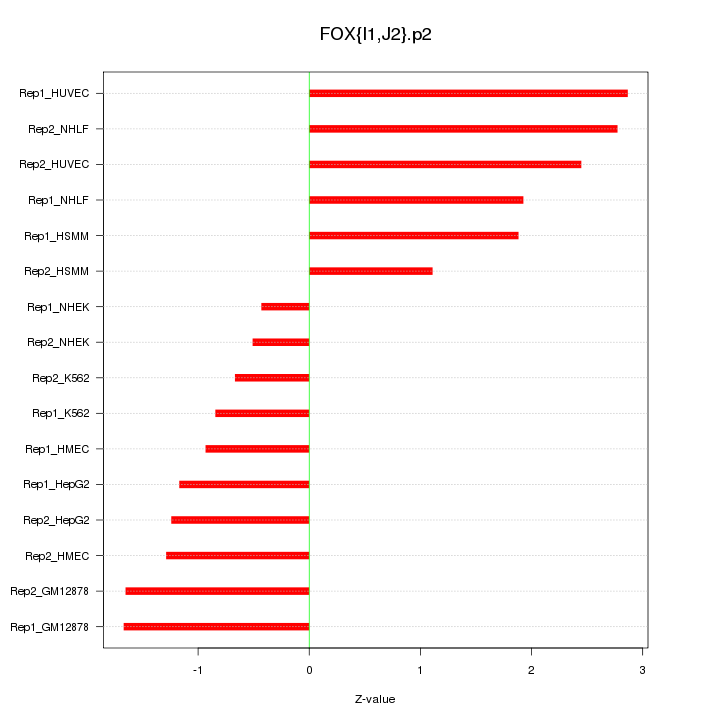
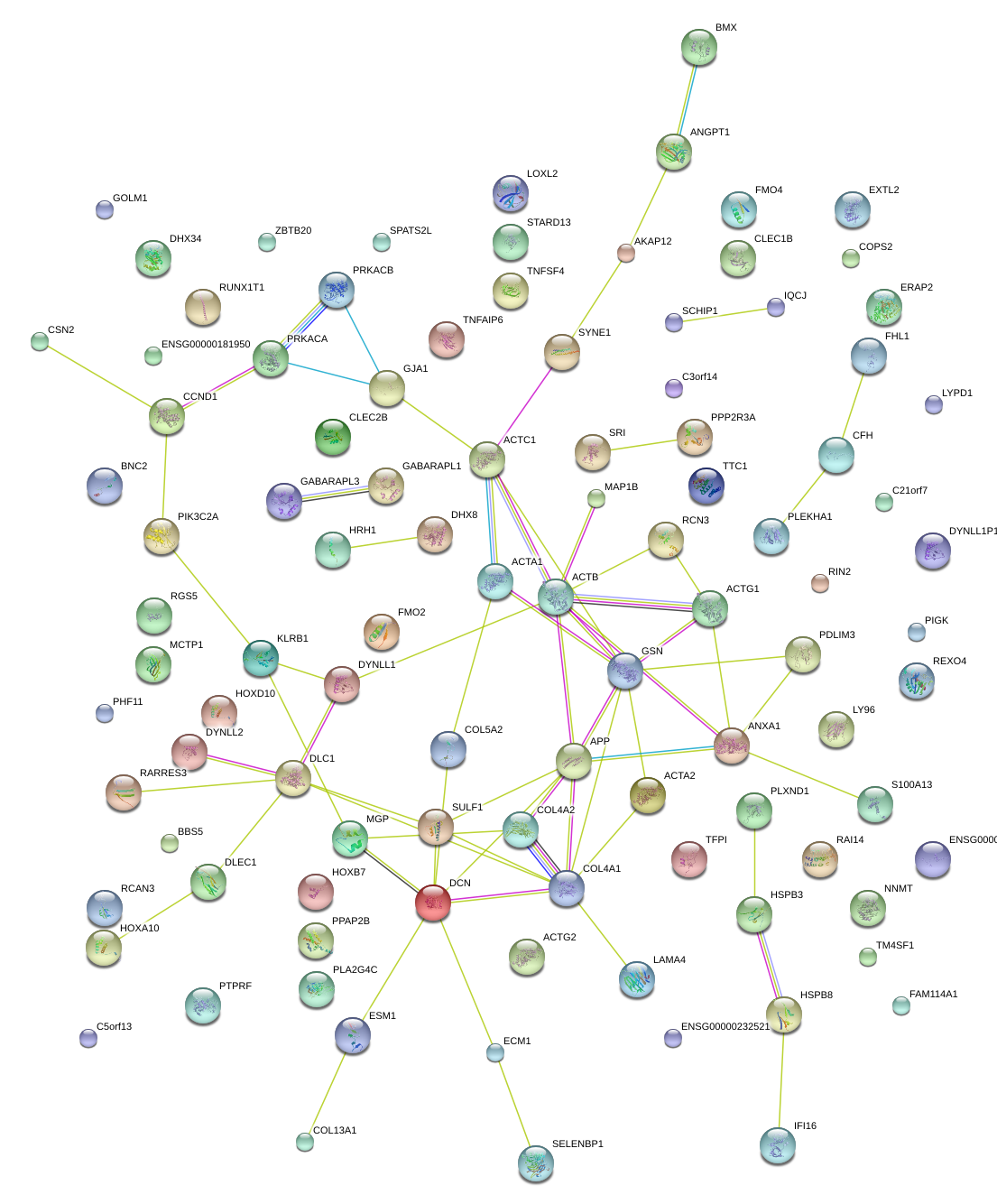
|
chr20_+_19818209
|
8.160
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_+_148747110
|
6.141
|
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr3_+_160269734
|
4.592
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr9_+_74956601
|
4.250
|
|
ANXA1
|
annexin A1
|
|
chr9_+_74956494
|
3.913
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr20_+_19863716
|
3.903
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr5_+_159368690
|
3.864
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr9_+_123123364
|
3.791
|
|
GSN
|
gelsolin
|
|
chr12_-_90100679
|
3.708
|
|
DCN
|
decorin
|
|
chr5_+_159368721
|
3.655
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr2_-_189752575
|
3.266
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr1_-_77457664
|
3.143
|
NM_005482
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K
|
|
chr5_+_159368733
|
3.106
|
NM_003314
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr13_-_109600709
|
3.074
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr9_-_87904844
|
3.030
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr12_-_90100733
|
2.912
|
NM_001920
|
DCN
|
decorin
|
|
chr11_+_113672378
|
2.704
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr3_-_150569574
|
2.608
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr11_+_113672407
|
2.600
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chrX_+_135106578
|
2.583
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_-_56817823
|
2.548
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr6_+_121798443
|
2.441
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr1_-_56817802
|
2.397
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr13_+_109932934
|
2.382
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr1_+_84382529
|
2.359
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr9_+_74956614
|
2.344
|
|
ANXA1
|
annexin A1
|
|
chr2_+_200878848
|
2.328
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr3_-_130761967
|
2.292
|
|
PLXND1
|
plexin D1
|
|
chr1_-_151866367
|
2.288
|
NM_001024212
|
S100A13
|
S100 calcium binding protein A13
|
|
chr1_-_151866147
|
2.243
|
NM_001024213
|
S100A13
|
S100 calcium binding protein A13
|
|
chr5_-_111340421
|
2.230
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_53787187
|
2.202
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr13_+_48968097
|
2.155
|
|
PHF11
|
PHD finger protein 11
|
|
chr11_+_69176391
|
2.145
|
|
CCND1
|
cyclin D1
|
|
chr5_-_54317102
|
2.027
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr5_+_71538456
|
2.004
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr6_+_151688327
|
2.000
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr7_-_87687276
|
1.960
|
|
SRI
|
sorcin
|
|
chr6_-_112682573
|
1.918
|
|
LAMA4
|
laminin, alpha 4
|
|
chr8_-_93181091
|
1.913
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_137224266
|
1.907
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr6_-_112682459
|
1.892
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr6_-_152681171
|
1.847
|
NM_015293
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr1_-_149611768
|
1.843
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr2_+_200879040
|
1.817
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr1_+_169421011
|
1.796
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr19_-_53305825
|
1.794
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr12_-_14929991
|
1.776
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr12_-_9913199
|
1.770
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr3_+_62280435
|
1.753
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr5_+_34723420
|
1.750
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr4_+_38545741
|
1.734
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr10_+_71231649
|
1.715
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr10_+_71231693
|
1.682
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr6_-_112682401
|
1.665
|
|
LAMA4
|
laminin, alpha 4
|
|
chr13_-_32757835
|
1.636
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr8_-_108579270
|
1.615
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr10_-_90702490
|
1.568
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr2_-_188127430
|
1.556
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_+_151922350
|
1.551
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr3_+_11242666
|
1.545
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr7_-_27186400
|
1.527
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr2_-_133145539
|
1.517
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr12_-_90100710
|
1.497
|
|
DCN
|
decorin
|
|
chr11_+_63060848
|
1.480
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr1_-_171441234
|
1.454
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr8_+_75066117
|
1.452
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr8_-_93176618
|
1.431
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_157246305
|
1.383
|
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr7_-_87687302
|
1.370
|
NM_003130
|
SRI
|
sorcin
|
|
chr21_+_29439768
|
1.341
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr12_+_118100977
|
1.337
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr5_-_94250357
|
1.331
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr17_-_44043361
|
1.299
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr2_+_170044194
|
1.297
|
NM_152384
|
BBS5
|
Bardet-Biedl syndrome 5
|
|
chr8_-_13018123
|
1.286
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_-_161439487
|
1.282
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_+_84402962
|
1.275
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr5_-_111119846
|
1.264
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_+_124135574
|
1.258
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr1_-_56817717
|
1.251
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_+_96237923
|
1.233
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr11_-_17147863
|
1.219
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr3_-_115825742
|
1.213
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr8_-_23317620
|
1.211
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr4_-_186693574
|
1.199
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr1_-_101132993
|
1.196
|
NM_001033025
NM_001439
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chrX_+_15428820
|
1.185
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr8_-_23317577
|
1.185
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr8_+_70567581
|
1.184
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr8_-_13178399
|
1.180
|
|
DLC1
|
deleted in liver cancer 1
|
|
chr19_+_54723044
|
1.169
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr9_-_16717887
|
1.163
|
|
BNC2
|
basonuclin 2
|
|
chr8_-_93177057
|
1.150
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_43837096
|
1.147
|
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr4_-_70861301
|
1.141
|
NM_001891
|
CSN2
|
casein beta
|
|
chr12_+_10256682
|
1.115
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr16_+_8622974
|
1.111
|
NM_024109
|
METTL22
|
methyltransferase like 22
|
|
chr21_-_26175068
|
1.105
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr2_+_201182187
|
1.105
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr1_+_99884018
|
1.102
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr1_+_15637521
|
1.095
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr12_-_9913724
|
1.084
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr5_+_15553288
|
1.082
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr1_+_19842309
|
1.076
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr4_+_15313670
|
1.073
|
NM_004334
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr1_+_221966684
|
1.055
|
NM_001748
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr17_-_45617899
|
1.049
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr2_+_102320148
|
1.042
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr1_-_103269356
|
1.035
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr10_-_126839056
|
1.009
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr3_+_185375362
|
0.993
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr10_+_53744043
|
0.990
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr3_+_185375356
|
0.985
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr11_-_104332631
|
0.973
|
NM_033306
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr2_-_188127294
|
0.972
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr20_+_42644633
|
0.971
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr10_-_126839000
|
0.968
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr6_+_153060722
|
0.965
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr4_-_16509446
|
0.960
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr3_+_185375370
|
0.954
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr4_-_16509354
|
0.948
|
|
LDB2
|
LIM domain binding 2
|
|
chr20_-_43972061
|
0.944
|
|
PLTP
|
phospholipid transfer protein
|
|
chr17_-_45619038
|
0.943
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr3_+_45042754
|
0.938
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr9_+_132002692
|
0.938
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr1_+_82038669
|
0.937
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr2_+_101874765
|
0.937
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr9_-_116920232
|
0.929
|
NM_002160
|
TNC
|
tenascin C
|
|
chr7_-_22506320
|
0.928
|
NM_001164460
NM_207342
|
MGC87042
|
STEAP family protein MGC87042
|
|
chr2_-_151052391
|
0.922
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr15_+_22619886
|
0.920
|
NM_022806
NM_022807
NM_022808
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chrX_-_10761729
|
0.920
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr2_-_179380301
|
0.917
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr11_-_27679617
|
0.913
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr20_+_9236446
|
0.913
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr8_-_17702665
|
0.912
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr3_+_185375379
|
0.906
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr7_+_89621624
|
0.903
|
NM_012449
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1
|
|
chr3_-_15357878
|
0.901
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr7_-_143213296
|
0.899
|
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr9_-_14076901
|
0.890
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_169326641
|
0.890
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr8_+_144711619
|
0.881
|
NM_024736
|
GSDMD
|
gasdermin D
|
|
chr12_+_29193399
|
0.877
|
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr4_-_16509126
|
0.873
|
|
LDB2
|
LIM domain binding 2
|
|
chr22_-_34754343
|
0.870
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr4_-_91979113
|
0.867
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr19_-_4351470
|
0.867
|
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr22_-_41366550
|
0.866
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr2_+_20510312
|
0.862
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr4_+_91035025
|
0.861
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr3_+_185375263
|
0.860
|
NM_001025205
NM_004068
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr10_+_63331018
|
0.857
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr7_+_89621725
|
0.856
|
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1
|
|
chr12_+_76749199
|
0.847
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr1_+_162795328
|
0.841
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr1_-_110735158
|
0.829
|
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr1_-_20682541
|
0.825
|
|
|
|
|
chr9_+_123126652
|
0.825
|
|
GSN
|
gelsolin
|
|
chr19_-_41215610
|
0.817
|
NM_015526
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr4_+_26194643
|
0.813
|
NM_018317
|
TBC1D19
|
TBC1 domain family, member 19
|
|
chr3_-_170347019
|
0.812
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr7_+_6583560
|
0.810
|
NM_001134387
NM_001134388
NM_001134389
NM_018106
|
ZDHHC4
|
zinc finger, DHHC-type containing 4
|
|
chr4_+_146623405
|
0.808
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr17_-_44043278
|
0.807
|
|
HOXB7
|
homeobox B7
|
|
chr18_+_71051713
|
0.799
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr4_-_5071996
|
0.799
|
NM_018659
|
CYTL1
|
cytokine-like 1
|
|
chr12_-_78831821
|
0.792
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory (inhibitor) subunit 12A
|
|
chr2_-_188021117
|
0.786
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr1_+_156229686
|
0.778
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr22_-_34566367
|
0.760
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_-_128883125
|
0.760
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr1_+_78243181
|
0.758
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr15_+_62467055
|
0.753
|
NM_016213
|
TRIP4
|
thyroid hormone receptor interactor 4
|
|
chr19_+_54722686
|
0.744
|
NM_020650
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr8_+_70567634
|
0.743
|
|
SULF1
|
sulfatase 1
|
|
chr7_+_75864755
|
0.741
|
NM_007155
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor)
|
|
chr11_+_65414677
|
0.739
|
|
CCDC85B
|
coiled-coil domain containing 85B
|
|
chr18_+_20260603
|
0.735
|
NM_018439
|
IMPACT
|
Impact homolog (mouse)
|
|
chr19_-_56946911
|
0.735
|
NM_001193306
NM_002029
|
FPR1
|
formyl peptide receptor 1
|
|
chr21_-_45118037
|
0.729
|
NM_004339
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr18_+_31488530
|
0.727
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr13_+_48967746
|
0.712
|
NM_001040443
|
PHF11
|
PHD finger protein 11
|
|
chr1_-_168310232
|
0.711
|
NM_014970
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr11_-_8695974
|
0.711
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr18_+_20260609
|
0.708
|
|
IMPACT
|
Impact homolog (mouse)
|
|
chr3_-_170346760
|
0.705
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr7_-_94902149
|
0.700
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr3_+_185375341
|
0.699
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr1_+_163084934
|
0.694
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr7_+_115926679
|
0.692
|
NM_001233
NM_198212
|
CAV2
|
caveolin 2
|
|
chr10_-_29794484
|
0.689
|
|
SVIL
|
supervillin
|
|
chr1_-_56817568
|
0.684
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_+_183290912
|
0.684
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr3_-_69120188
|
0.676
|
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr4_+_41056378
|
0.675
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_124537334
|
0.673
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_+_143807748
|
0.668
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr10_-_104252394
|
0.665
|
NM_005736
|
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast)
|